Genomic Selection

Knowing the strengths and weaknesses of young cattle already when the animals are mated for the first time (Photo: Han Hopman)
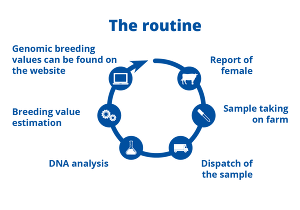
While until 2010 the entire selection of young cattle had to be done via the pedigree index (i.e. based on parental breeding values), genomic breeding values today offer a more targeted look at the genetic potential of the next generation. In order to estimate genomic breeding values, genotyping must first be carried out in the laboratory.
In genotyping, the DNA of a living organism is analysed for a large number of selected markers. These markers are called SNPs (single nucleotide polymorphisms). SNPs are hereditary point mutations and therefore occur as a swapped base in the DNA strand. Usually, about 50,000 SNPs are examined in the laboratory at the same time. The laboratory results can occur in three combinations for each SNP (twice allele 1 (homozygous), once allele 1 and once allele 2 (heterozygous) or twice allele 2 (homozygous)). A SNP rarely occurs directly within a gene and, for example, forms the causal mutation for different trait expressions (phenotypes). Nevertheless, SNPs have a high informative value if they are located close to important genes.

Already a few days after birth, a tissue sampling tag is used to get a small amount of tissue from a calf for genotyping (Photo: D. Warder)
To genotype an animal, the farmer sends a sample with genetic material (e.g. blood, tissue or hair) and an application for testing to an accredited laboratory. Once there, the DNA is extracted from the sample. It is then fragmented and amplified. The DNA fragments are applied to a chip which contains a kind of docking site for the DNA fragments. Depending on the expression of a SNP, the strand connects to one of the possible docking sites. Through a light signal, which is emitted by a marked base at the docking site when excited by a laser, the information about the SNP can then be identified by the computer. Accordingly, there are also three colour signals for the three possible expressions of a SNP. Afterwards, the laboratory sends the typing results to the data centre in Verden (vit). The information is then processed there and genomic breeding values are estimated.
The breeding value estimation takes into account the SNP effects. This means that the closer a SNP is to a gene (with an influence on a trait under consideration), the greater is its effect in the breeding value estimation model towards that particular trait. Of course, this requires that the gene and the SNP occur in the same possible expressions. In this case, it is called a linkage. The genomic breeding values are then provided to the breeding organisations and the owners of the animals via an online platform.
Farmers and breeding organisations are now in a position to make specific selection decisions at a very young age based on genomic breeding values.
KuhVision and herd genotyping
In Germany, the genotyping of female animals of the breed German Holsteins is available for all herdbook farms. In principle, it is possible to apply for genotyping of individual animals at any time. However, the entire female offspring can also be typed after birth. For this purpose, farms sign a contract with their breeding organisation and receive a discount on the typing costs. Farmers who decide to additionally record health data can receive a further discount in the KuhVision project.
In the meantime, about 20% of herdbook animals are genotyped via this system and thus make a valuable contribution to improving genomic breeding value estimation.
Participation in these projects results in a variety of benefits for the participants:
- Herd genotyping offers a precise overview of the genetic status of the herd through genomic breeding values with a high degree of reliability.
- Breeding values for direct health traits can be used as an indicator of the genetic susceptibility of economically significant disease complexes.
- By selecting the genetically best animals, the number of inseminations required for herd replacement can be reduced.
- A selective use of sexed semen additionally increases the breeding progress in the herd.
- Offspring with a lower genetic level can be sold as calves and therefore save rearing costs and resources.
- Cows that are not needed for herd replacement can be sold or mated with beef breeds.
- Genetic weaknesses, e.g. in animal health, can be improved more effectively. For example, a reduction in digital dermatitis can be enhanced by using bulls with a genetically lower susceptibility.
- By using the Bull Breeding Programme (BAP), individual breeding goals can be set and pursued.
- The information on the genetic horn status makes it easier to breed polled animals.
- Computer-assisted parentage control of the genotyped animals improves the accuracy of the pedigrees.
- The risk of mating with genetic defect carriers can be reduced.
- Genomic information allows the reduction / limitation of the degree of inbreeding in the herd.
- Farms can be compared at the level of breeding values.
- Breeding values are published once a week and therefore results will be available within two weeks after sending a sample.
The genotyping of male calves and bulls is only done within the breeding programmes for the selection of possible new breeding bulls.
Further information can be found here and on the websites of the German breeding organisations.